News Story
BIOE, IBBR Researchers Develop Electrogenetic Device for Activating Gene Expression via Electrodes

Researchers from the University of Maryland (UMD) are working to develop an electrogenetic device to direct gene expression, an achievement that holds promise for controlling biological systems and could help shape the future of biosensors, as well as wearable – and possibly implantable – bio-hybrid devices.
The team, led by William Bentley, UMD Distinguished University Professor and director of the university’s Robert E. Fischell Institute for Biomedical Devices, is using redox biomolecules – tiny cellular messengers that are vital to the health of all body cells – to carry electronic information to engineered bacterial cells. To do this, the group has developed a patent-pending electrogenetic device that uses an electrode and engineered cells to control how and when genes are expressed from a synthetic gene circuit.
“Researchers have long used microelectronic devices embedded with biological components, such as high throughput DNA sequencing technologies, to interrogate biology, but such devices have the potential to do much more – perhaps even allow scientists to control biology,” Bentley said.
Previously, Bentley’s team took a first step by developing a methodology to load and control enzymes onto microelectronic chips by modulating natural redox molecules located between the enzymes and the electrode. Building on this, the team is now modulating redox molecules in order to link electrode-actuated signals to cells specifically engineered to respond by activating gene expression. This methodology could open doors for scientists looking to drive intricate biological behaviors – such as by controlling biofilms or even producing therapeutics in microdevices.
Expanding on previous applications of electrogenetic devices in synthetic biology, Bentley, along with fellow Fischell Department of Bioengineering (BIOE) and UMD Institute for Bioscience and Biotechnology Research (IBBR) professor Gregory Payne, and the research team have demonstrated that small molecules offer a wide repertoire for molecular communication. As noted in the group’s Nature Communications paper published online Tuesday, Jan. 17, once scientists gain the ability to measure, disrupt, or enhance these biomolecular signals, they will be well-positioned to develop advanced technologies to study and manipulate the biological environment.
“Electronics have transformed the way we live our lives, and there have been increasing efforts to ‘connect’ devices to biology, such as with glucometers or fitness trackers that access biological information,” Payne said. “But, there are far fewer examples of electronics communicating in the other direction to provide the cues that guide biological responses. Such capabilities could offer the potential to apply devices to better fight diseases such as cancer or to guide inflammatory responses to promote wound healing.”
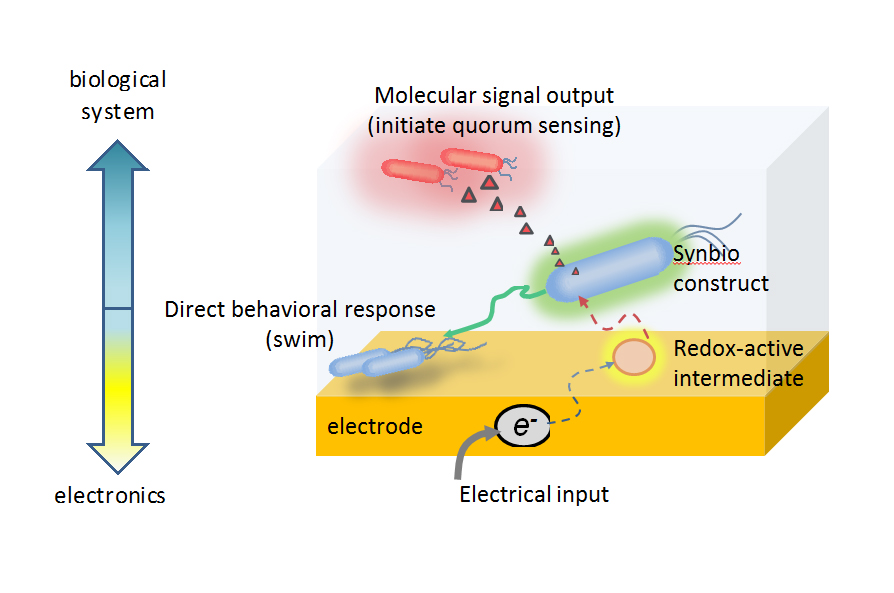 Applying their methodology, the team demonstrated the ability to control gene expression to induce the movement of bacteria – known as “bacterial swimming” – and to build a cellular information relay in which one group of bacteria interprets the electronic signal and passes the information to another group to change its gene expression.
Applying their methodology, the team demonstrated the ability to control gene expression to induce the movement of bacteria – known as “bacterial swimming” – and to build a cellular information relay in which one group of bacteria interprets the electronic signal and passes the information to another group to change its gene expression.
The team’s circuit relies on redox signaling processes prevalent in most biological systems, including the human body. Redox processes are involved in protecting the body from oxidative damage, such as when a person is exposed to bright sunlight.
“Like our bodies, bacteria have adapted ways to escape oxidative damage,” Bentley said. “Our team has engineered bacterial cells to instead interpret the oxygen signaling processes, and we have developed a genetic circuit that relies on the associated molecular cues for actuating a programmed response.”
To guide the production of proteins that direct cell function, Bentley and his team – which includes the paper’s first author, IBBR researcher and BIOE alumna Tanya Tschirhart (Ph.D. ’15) – tap into this process by using pyocyanin (Pyo), a metabolite and molecular signal with the ability to oxidize and reduce other molecules for gene induction. To utilize Pyo in this way, Bentley’s team is working with a redox-responsive regulon in E.coli to sense and respond to oxidative stress. This enables the team to turn “on” or “off” the gene expression or, specifically, protein production of the expressed gene.
Even more, by electronically controlling the oxidation state of another redox molecule, ferricyanide, Bentley’s team has been able to amplify the specific increases or decreases in protein levels in E. coli. Their efforts have demonstrated for the first time the utility of using biologically relevant redox molecules in translating electronic signals to changes in engineered bacterial gene expression. Further, they have showed that electronically activated cells could be made to send natural, biological signal molecules to neighboring cells, to ultimately control their behavior. In this way, the group’s electrogenetic device can be “programmed” to control “remote” biological behavior.
Unlike previous synthetic biology efforts, Bentley’s group carried out minimal “rewiring” of cells to take advantage of native redox interactions and provide insights into their developing role as mediators for bioelectronic communication.
The group believes their system can be tailored to produce a variety of responses, guide various cell behaviors, and further the use of other electronic and redox-based systems to access and affect biomolecular information transfer, such as in microbial fuel cells or bioelectrosynthesis systems. Additionally, the team’s electrogenetic device could serve as a powerful addition to the “biofabrication” toolbox, furthering the utilization of biologically-inspired nanoscale processes by bridging the fabrication and communication gaps between microelectronics and biological systems.
In addition to Bentley, Payne, and Tschirhart, research team members and co-authors include: Eunkyoung Kim (IBBR), Ryan McKay (IBBR, BIOE), Hana Ueda (IBBR, UMD Department of Mathematics), Hsuan-Chen Wu (IBBR), Alex (Eli) Pottash (IBBR, BIOE), and Amin Zargar (BIOE), as well as Alejandro Negrete and Joseph Shiloach of the National Institutes of Health’s National Institutes of Diabetes and Digestive and Kidney Diseases Biotechnology Core Laboratory.
This research is supported by the Defense Threat Reduction Agency and the National Science Foundation.
Full text of the Nature Communications paper, “Electronic control of gene expression and cell behavior in Escherichia coli through redox signaling,” is available online.
Published January 17, 2017